Protein analysis web servers by the group, freely accesible for academic users. They include: Pirepred, Protposer, ProtSA, PrionScan and ProteinLIPS
01. PirePred
PirePred is a genetic interpretation tool to suit the needs of Newborn Screening Programs. Missense, nonsense and frameshift single nucleotide variants (SNVs) annotated in the ClinVar database for 58 genes related to newborn screening are retrieved in real time and presented in the structural context of the original protein. For each variant, binary classifications (tolerated/deleterious) obtained from 16 popular predictors are shown, together with a consensus ternary classification (tolerated/uncertain/deleterious). Patogenicity predictions for any single amino acid replacement arising from SNVs in those genes, whether already anotated in ClinVar or not, are also shown.


02. Protposer
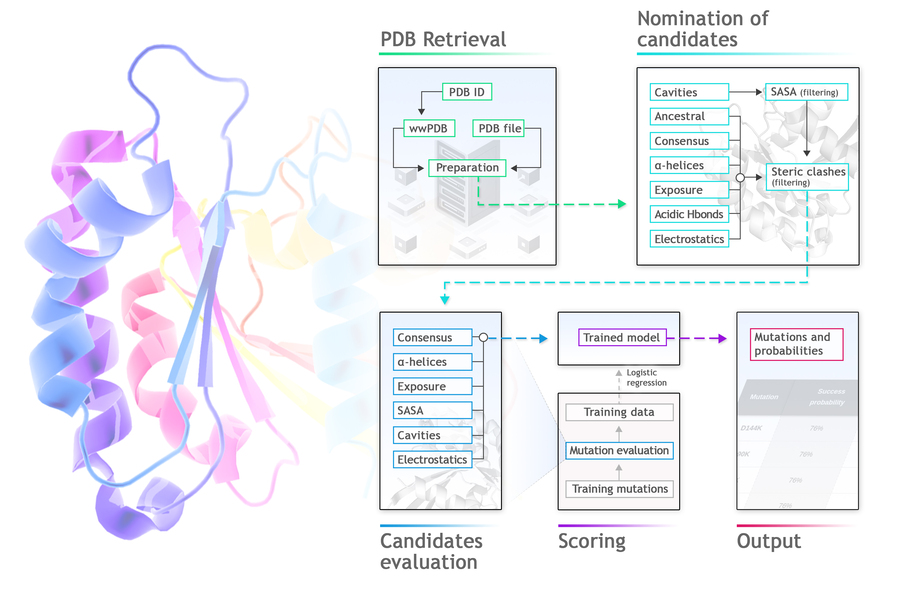
Protposer is a protein stabilization tool that analyses PDB files and sequence databases in search for wild type amino acid residues providing opportunities for thermostabilization. Protposer proposes a list of point mutations that are likely to increase protein stability by at least 0.5 kcal/mol. Protposer finds weak spots in the structure, generates and evaluates replacements, and estimates their probability of being stabilizing, using a machine learning logistic regression model.To do so, it searches for divergences from ancestral or consensus sequences, from ideal alpha-helix composition or average solvent exposure of polar or apolar residues, and for the presence of cavities, electrostatic imbalance or acidic H-bonds.
03. ProtSA
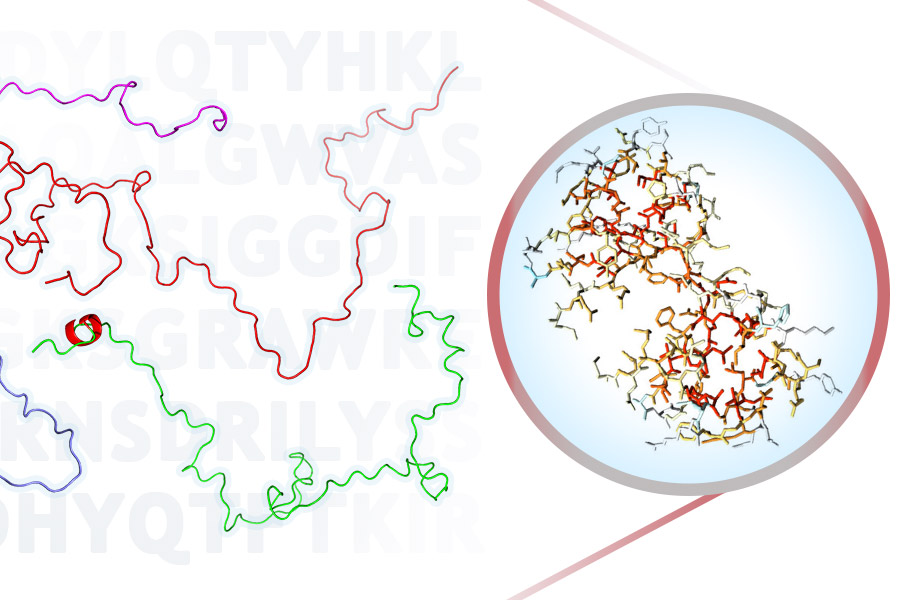
ProtSA is a tool to model and analyse the unfolded ensemble of a protein. From the sequence of your protein of interest, ProtSA generates an unfolded ensemble of upto 2000 different conformations (which you may download as PDB files), and it calculates the average solvent accessible surface area (SASA) of each atom and residue in the ensemble. In addition, if you provide a PDB file of the folded state, folded SASA and differences between the folded and unfolded SASA, per atom and per residue, will also be returned.

04. PrionSCAN
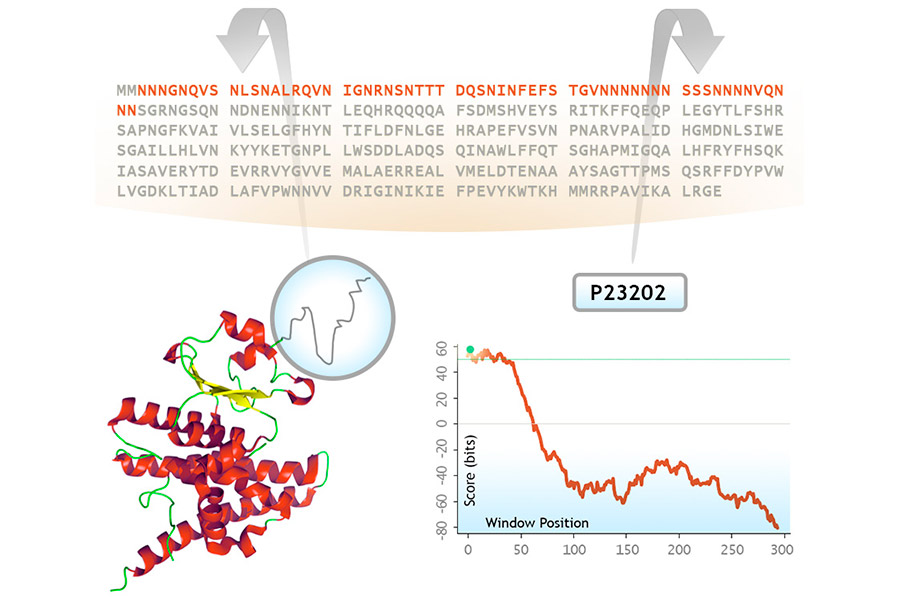
PrionSCAN predicts prion-like sequence stretches in the proteomes annotated in UniprotKB. Based on amino acid propensities previously observed in prion domains experimentally tested in yeast, we have built a probabilistic model, benchmarked to handle large sequence databases, and used it to predict prion-like sequence stretches in proteins.

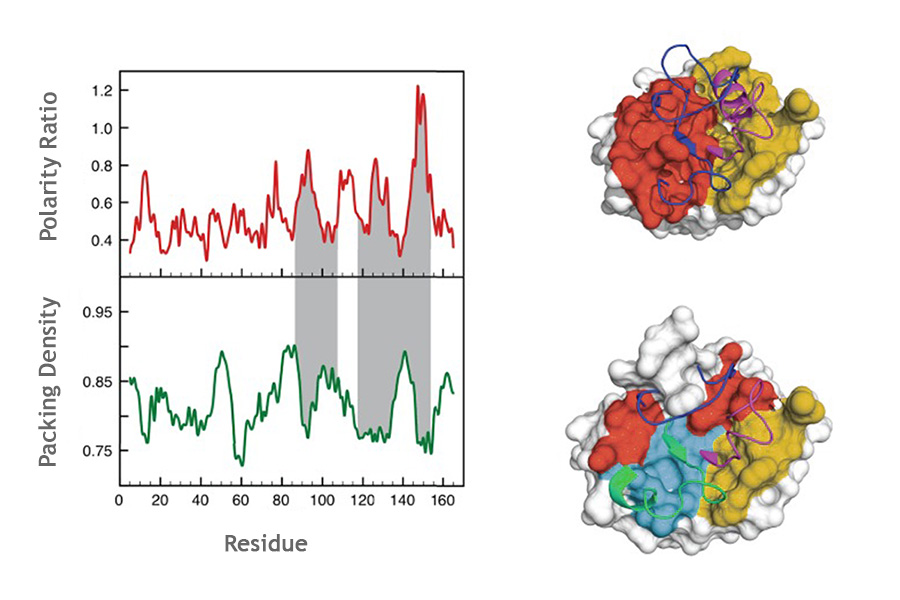
05. ProteinLIPS
ProteinLIPS identify unstable regions of non-fully cooperative protein domain. It scans protein structures, computes packing densities and surface polarities, and predicts the locally unstable segments which are likely to be the first parts of the protein to unfold under thermal stress, the last parts to fold, or to be highly dynamic.

